I am currently doing exercises from digital house brasil
Libraries
Let’s see what version of python this env is running.
import platform
print(platform.python_version())## 3.6.12import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import os
file_path_linux = r.file_path_linuxThe Exercise
Before we get into it
Objectives
Open and read a DataFrame using pandas
Basic analysis of each column using value counts.
Creating a hypothesis that we care about
In our case the hypothesis is simple do women earn on average less than men?
Data preprocessing
To do in the second post
Define the variables used in the conclusion
In our case, we choose to use salary ~ sex,region region was added to test whether Simpson’s paradox was at play.
Using masks or other methods to filter the data
This objective was mostly done using the groupby function.
Visualizing the hypothesis
We were advised to use two histograms combined to get a preview of our answer.
Conclusion
Comment on our findings.
Reservations
This is an exercise where we were supposed to ask a relevant question using the data from the IBGE(Brazil’s main data collector) database of 1970.
Our group decided to ask whether women received less than man, we expanded the analysis hoping to avoid the Simpson’s paradox.
This is just an basic inference, and it’s results are therefore only used for studying purposes I don’t believe any finding would be relevant using just this approach but some basic operations can be used in a more impact full work.
Data Dictionary
We got a Data Dictionary that will be very useful for our Analysis, it contains all the required information about the encoding of the columns and the intended format that the folks at STATA desired.
Portuguese
Descrição do Registro de Indivíduos nos EUA.
Dataset do software STATA (pago), vamos abri-lo com o pandas e transforma-lo em DataFrame.
Variável 1 – CHAVE DO INDIVÍDUO ? Formato N - Numérico ? Tamanho 11 dígitos (11 bytes) ? Descrição Sumária Identifica unicamente o indivíduo na amostra.
Variável 2 - IDADE CALCULADA EM ANOS ? Formato N - Numérico ? Tamanho 3 dígitos (3 bytes) ? Descrição Sumária Identifica a idade do morador em anos completos.
Variável 3 – SEXO ? Formato N - Numérico ? Tamanho 1 dígito (1 byte) ? Quantidade de Categorias 3 ? Descrição Sumária Identifica o sexo do morador. Categorias (1) homem, (2) mulher e (3) gestante.
Variável 4 – ANOS DE ESTUDO ? Formato N - Numérico ? Tamanho 2 dígitos (2 bytes) ? Quantidade de Categorias 11 ? Descrição Sumária Identifica o número de anos de estudo do morador. Categorias (05) Cinco ou menos, (06) Seis, (07) Sete, (08) Oito, (09) Nove, (10) Dez, (11) Onze, (12) Doze, (13) Treze, (14) Quatorze, (15) Quinze ou mais.
Variável 5 – COR OU RAÇA ? Formato N - Numérico ? Tamanho 2 dígitos (2 bytes) ? Quantidade de Categorias 6 ? Descrição Sumária Identifica a Cor ou Raça declarada pelo morador. Categorias (01) Branca, (02) Preta, (03) Amarela, (04) Parda, (05) Indígena e (09) Não Sabe.
Variável 6 – VALOR DO SALÁRIO (ANUALIZADO) ? Formato N - Numérico ? Tamanho 8 dígitos (8 bytes) ? Quantidade de Decimais 2 ? Descrição Sumária Identifica o valor resultante do salário anual do indivíduo. Categorias especiais (-1) indivíduo ausente na data da pesquisa e (999999) indivíduo não quis responder.
Variável 7 – ESTADO CIVIL ? Formato N - Numérico ? Tamanho 1 dígito (1 byte) ? Quantidade de Categorias 2 ? Descrição Sumária Dummy que identifica o estado civil declarado pelo morador. Categorias (1) Casado, (0) não casado.
Variável 8 – REGIÃO GEOGRÁFICA ? Formato N - Numérico ? Tamanho 1 dígito (1 byte) ? Quantidade de Categorias 5 ? Descrição Sumária Identifica a região geográfica do morador. Categorias (1) Norte, (2) Nordeste, (3) Sudeste, (4) Sul e (5) Centro-oeste.
English
Description of the US Individual Registry.
Dataset of the STATA software (paid), we will open it with pandas and turn it into DataFrame.
Variable 1 - KEY OF THE INDIVIDUAL? Format N - Numeric? Size 11 digits (11 bytes)? Summary Description Uniquely identifies the individual in the sample.
Variable 2 - AGE CALCULATED IN YEARS? Format N - Numeric? Size 3 digits (3 bytes)? Summary Description Identifies the age of the resident in full years.
Variable 3 - SEX? Format N - Numeric? Size 1 digit (1 byte)? Number of Categories 3? Summary Description Identifies the gender of the resident. Categories (1) men, (2) women and (3) pregnant women.
Variable 4 - YEARS OF STUDY? Format N - Numeric? Size 2 digits (2 bytes)? Number of Categories 11? Summary Description Identifies the number of years of study of the resident. Categories (05) Five or less, (06) Six, (07) Seven, (08) Eight, (09) Nine, (10) Dec, (11) Eleven, (12) Twelve, (13) Thirteen, (14 ) Fourteen, (15) Fifteen or more.
Variable 5 - COLOR OR RACE? Format N - Numeric? Size 2 digits (2 bytes)? Number of Categories 6? Summary Description Identifies the Color or Race declared by the resident. Categories (01) White, (02) Black, (03) Yellow, (04) Brown, (05) Indigenous and (09) Don’t know.
Variable 6 - WAGE VALUE (ANNUALIZED)? Format N - Numeric? Size 8 digits (8 bytes)? Number of decimals 2? Summary Description Identifies the amount resulting from the individual’s annual salary. Special categories (-1) individual absent on the survey date and (999999) individual did not want to answer.
Variable 7 - CIVIL STATE? Format N - Numeric? Size 1 digit (1 byte)? Number of Categories 2? Summary Description Dummy that identifies the marital status declared by the resident. Categories (1) Married, (0) Not married.
Variable 8 - GEOGRAPHICAL REGION? Format N - Numeric? Size 1 digit (1 byte)? Number of Categories 5? Summary Description Identifies the resident’s geographic region. Categories (1) North, (2) Northeast, (3) Southeast, (4) South and (5) Midwest.
Python
Pre-processing
Reading Data
The path is specific for my computer but it is easy to adapt
You can also dowload it from the github page from this blog
# Abertura e leitura dos dados em um DeteFrame em Pandas
path = r.file_path_linux
df = pd.read_csv(path + '/stata_data_1970.csv')Analyzing some basic stuff about our data frame
#Análise básica dos conteúdos de cada coluna com contagem de valores
df.info()## <class 'pandas.core.frame.DataFrame'>
## RangeIndex: 66470 entries, 0 to 66469
## Data columns (total 9 columns):
## # Column Non-Null Count Dtype
## --- ------ -------------- -----
## 0 Unnamed: 0 66470 non-null int64
## 1 id 66470 non-null float64
## 2 idade 66470 non-null int64
## 3 sexo 66470 non-null object
## 4 anos_estudo 66036 non-null float64
## 5 cor/raca 66228 non-null object
## 6 salario 47878 non-null float64
## 7 estado_civil 66470 non-null float64
## 8 regiao 66470 non-null object
## dtypes: float64(4), int64(2), object(3)
## memory usage: 4.6+ MBI do enjoy python’s base value_counts but when used in a loop it can create some ugly outputs, in order to fix I created a function that adds some flavor text to the print output and generates new information about the accumulated percentage of the data being displayed.
Custom count_values()
def pretty_value_counts(data_frame,
number_of_rows = 5,
cum_perc = True):
for col in data_frame:
counts = data_frame[col].value_counts(dropna=False)
percentages = data_frame[col].value_counts(dropna=False, normalize=True)
if cum_perc == True:
cum_percentages = percentages.cumsum()
tb = pd.concat([counts,
percentages,
cum_percentages],
axis=1,
keys=['counts',
'percentages',
"cum_percentages"]
).head(number_of_rows)
else:
tb = pd.concat([counts,
percentages],
axis=1,
keys=['counts',
'percentages']).head(number_of_rows)
print("Column %s with %s data type" % (col,data_frame[col].dtype),
"\n",
tb,
"\n")Now we can apply our new function.
Using a custom function
pretty_value_counts(df)## Column Unnamed: 0 with int64 data type
## counts percentages cum_percentages
## 2047 1 0.000015 0.000015
## 41601 1 0.000015 0.000030
## 21151 1 0.000015 0.000045
## 23198 1 0.000015 0.000060
## 17053 1 0.000015 0.000075
##
## Column id with float64 data type
## counts percentages cum_percentages
## 1.100351e+10 2 0.000030 0.000030
## 3.132701e+10 1 0.000015 0.000045
## 1.501501e+10 1 0.000015 0.000060
## 3.230631e+10 1 0.000015 0.000075
## 5.003991e+10 1 0.000015 0.000090
##
## Column idade with int64 data type
## counts percentages cum_percentages
## 20 2104 0.031653 0.031653
## 28 2056 0.030931 0.062585
## 26 2040 0.030691 0.093275
## 22 2034 0.030600 0.123875
## 27 2017 0.030345 0.154220
##
## Column sexo with object data type
## counts percentages cum_percentages
## mulher 33607 0.505597 0.505597
## homem 32791 0.493320 0.998917
## gestante 72 0.001083 1.000000
##
## Column anos_estudo with float64 data type
## counts percentages cum_percentages
## 5.0 23349 0.351271 0.351271
## 11.0 16790 0.252595 0.603866
## 15.0 5636 0.084790 0.688657
## 8.0 5017 0.075478 0.764134
## 10.0 2704 0.040680 0.804814
##
## Column cor/raca with object data type
## counts percentages cum_percentages
## Branca 31689 0.476741 0.476741
## Parda 28370 0.426809 0.903550
## Preta 5249 0.078968 0.982518
## Indigena 597 0.008981 0.991500
## Amarela 323 0.004859 0.996359
##
## Column salario with float64 data type
## counts percentages cum_percentages
## NaN 18592 0.279705 0.279705
## 0.0 1841 0.027697 0.307402
## -1.0 1101 0.016564 0.323966
## 999999.0 367 0.005521 0.329487
## 5229.0 277 0.004167 0.333654
##
## Column estado_civil with float64 data type
## counts percentages cum_percentages
## 1.0 39066 0.587724 0.587724
## 0.0 27404 0.412276 1.000000
##
## Column regiao with object data type
## counts percentages cum_percentages
## sudeste 25220 0.379419 0.379419
## centro-oeste 14702 0.221182 0.600602
## norte 14653 0.220445 0.821047
## sul 11890 0.178878 0.999925
## nordeste 5 0.000075 1.000000Just for comparison lets look how we could do the same thing without the function.
for col in df:
df[col].value_counts(dropna=False).head(5)## 2047 1
## 41601 1
## 21151 1
## 23198 1
## 17053 1
## Name: Unnamed: 0, dtype: int64
## 1.100351e+10 2
## 3.132701e+10 1
## 1.501501e+10 1
## 3.230631e+10 1
## 5.003991e+10 1
## Name: id, dtype: int64
## 20 2104
## 28 2056
## 26 2040
## 22 2034
## 27 2017
## Name: idade, dtype: int64
## mulher 33607
## homem 32791
## gestante 72
## Name: sexo, dtype: int64
## 5.0 23349
## 11.0 16790
## 15.0 5636
## 8.0 5017
## 10.0 2704
## Name: anos_estudo, dtype: int64
## Branca 31689
## Parda 28370
## Preta 5249
## Indigena 597
## Amarela 323
## Name: cor/raca, dtype: int64
## NaN 18592
## 0.0 1841
## -1.0 1101
## 999999.0 367
## 5229.0 277
## Name: salario, dtype: int64
## 1.0 39066
## 0.0 27404
## Name: estado_civil, dtype: int64
## sudeste 25220
## centro-oeste 14702
## norte 14653
## sul 11890
## nordeste 5
## Name: regiao, dtype: int64Replacing columns names
The columns are named in Portuguese we can replace their names for English equivalents in a lot of different ways
df.columns## Index(['Unnamed: 0', 'id', 'idade', 'sexo', 'anos_estudo', 'cor/raca',
## 'salario', 'estado_civil', 'regiao'],
## dtype='object')My favorite way of doing this sort of trades is using a dictionary defined outside the replace method, the cool thing about replace is that if we liked some of the column names previously defined we can simply omit them, for example, both “Unnamed: 0” and “id” are useless but since their names are already in English I don’t need to mess with them right now
Translation discussion on race
There is some valid discussion on whether to translate “cor/raca” into ethnic_group or color_race, but I am personally on the opinion that the ones making this data frame in 1970 were probably under other standards of naming conventions and racism accusations so I will keep their naming scheme, I apologize if anyone feels offended by the use of these terms
dict_cols = {"idade" : "age",
"sexo" : "sex",
"anos_estudo" : "years_study",
"cor/raca" : "color_race",
"salario" : "salary",
"estado_civil" : "civil_status",
"regiao" : "region"
}df.rename(columns = dict_cols, inplace = True)Let’s see what changed
df.columns## Index(['Unnamed: 0', 'id', 'age', 'sex', 'years_study', 'color_race', 'salary',
## 'civil_status', 'region'],
## dtype='object')It look fine now we can translate some of our main features
Cleaning categorical data
First we need to know the categories present in each of our columns a simple loop would fails us when we reached a numeric variable, the simplest way to solve that would be using an if statement, another alternative is using conditional execution, I personally don’t know a simple way of doing that in python but I will show it in the R post
To discover the numeric and “categorical” variables, know that sometimes you will have to change some elements of these lists but looking at my outputs I think I got all the relevant ones
Finding which columns are categorical
These are the numerical variables
df.select_dtypes(include=[np.number]).columns## Index(['Unnamed: 0', 'id', 'age', 'years_study', 'salary', 'civil_status'], dtype='object')And these are the Categorical variables
list_cat = df.select_dtypes(exclude=[np.number]).columnsNow we can run a simple loop
for col in list_cat:
df[col].unique()## array(['homem', 'mulher', 'gestante'], dtype=object)
## array(['Parda', 'Amarela', 'Indigena', 'Branca', 'Preta', nan],
## dtype=object)
## array(['norte', 'nordeste', 'sudeste', 'sul', 'centro-oeste'],
## dtype=object)The simpler method is comparing the dtype in each column to the desired output, but this would be harder if we needed the np.numeric
for col in df:
if df[col].dtype == "O":
df[col].unique()## array(['homem', 'mulher', 'gestante'], dtype=object)
## array(['Parda', 'Amarela', 'Indigena', 'Branca', 'Preta', nan],
## dtype=object)
## array(['norte', 'nordeste', 'sudeste', 'sul', 'centro-oeste'],
## dtype=object)The problem with the simpler approach is that sometimes you have columns that are categories and not objects so the simpler approach would fail when the more complex one would not, let’s convert sex to a category to prove my point
df.sex =df.sex.astype("category")df.dtypes## Unnamed: 0 int64
## id float64
## age int64
## sex category
## years_study float64
## color_race object
## salary float64
## civil_status float64
## region object
## dtype: objectfor col in df:
if df[col].dtype == "O":
df[col].unique()## array(['Parda', 'Amarela', 'Indigena', 'Branca', 'Preta', nan],
## dtype=object)
## array(['norte', 'nordeste', 'sudeste', 'sul', 'centro-oeste'],
## dtype=object)It does not work anymore, of course you can still solve this “problem” with the simpler approach by including a “and” clause on your if statement but at that point you might as well use the more extensible appoach
Replacing values with an dictionary: 1 column
After looking into the categories I can create a dictionary for each column if I want to be safe on repeating terms or I can pass a master dictionary for the whole data frame, I think the column by column approach is tidier but for each their own
dict_sex = {"mulher" : "woman",
"homem" : "man",
"gestante" : "woman"} # pregnantThis is one strange data frame, it probably made sense to split women into pregnant and not pregnant but I think it will only complicate the otherwise simple analyses so I will group both into “woman”
df.sex.replace(dict_sex,inplace = True)Showing the new amounts of women/mean
df.sex.value_counts()## woman 33679
## man 32791
## Name: sex, dtype: int64df.sex.unique()## array(['man', 'woman'], dtype=object)This fails
pretty_value_counts(df.sex)## Error in py_call_impl(callable, dots$args, dots$keywords): KeyError: 'man'
##
## Detailed traceback:
## File "<string>", line 1, in <module>
## File "<string>", line 6, in pretty_value_counts
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\series.py", line 882, in __getitem__
## return self._get_value(key)
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\series.py", line 990, in _get_value
## loc = self.index.get_loc(label)
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\indexes\range.py", line 358, in get_loc
## raise KeyError(key)Here is actually a example on why I don’t personally enjoy Pandas conversion of data, the function that we created pretty_value_counts is not gonna work in this example because Pandas converts a single column to an Series object, so we would have to write a pretty_value_counts for Series as well or we would have to mess with the Pandas method or we could convert the series back into a DataFrame like this
pretty_value_counts(pd.DataFrame(data= df.sex))## Column sex with object data type
## counts percentages cum_percentages
## woman 33679 0.50668 0.50668
## man 32791 0.49332 1.00000Replacing values with an dictionary: multiple columns
Translation discussion on race part 2
Again there is relevant discussion on whether I should translate “Parda” as brown but basically Brazil’s population sometimes answers that their skin color is “Parda” = brown when asked about for many reasons I will propose two, “Preta” black can be used as an racist term so some people prefer to be called “brown”, the second explanation is that most of the population is actually pretty well integrated meaning that there a lot of biracial couples in this case we see something like “Preta” parent + “Branca” parent = “Parda” = in English “brown”.
There is also the case for the English equivalent of brown skin we simply use “Indiano” = “Indian”.
Curiously the term “Negra” =~ "N*gger" is often preferred in Brazil, that may cause some confusion between Portuguese and English speakers.
I will use brown but do notice that there were multiple sensible approaches here.
This is a good opportunity to show failures in the master dictionary approach, realize that if I were to replace “nan” as no_answer or something like that python could thrown me an error because there are “nan” in some numerical columns such as salary but instead I get silence conversion of a numerical columns into object columns a dangerous feature.
for col in list_cat:
df[col].unique()## array(['man', 'woman'], dtype=object)
## array(['Parda', 'Amarela', 'Indigena', 'Branca', 'Preta', nan],
## dtype=object)
## array(['norte', 'nordeste', 'sudeste', 'sul', 'centro-oeste'],
## dtype=object)dict_all = {"Parda" : "brown",
"Amarela" : "yellow",
"Indigena" : "indigenous",
"Branca" : "white",
"Preta" : "black",
np.nan : "no_answer"}df.replace(dict_all).salary.dtype## dtype('O')dict_all = {"Parda" : "brown", #col color_race
"Amarela" : "yellow",
"Indigena" : "indigenous",
"Branca" : "white",
"Preta" : "black",
"norte" : "north", # col region
"nordeste" : "northeast",
"sudeste" : "southeast",
"sul" : "south",
"centro-oeste" : "midwest"}Let’s pray that we don’t have this problem and use this shared dictionary
df.replace(dict_all, inplace = True)Did we correctly clean the Categorical Variables?
Conversion of types
Well not really I would argue that year_study is an categorical variable as well so let’s convert it.
df.years_study = df.years_study.astype('category')df.years_study.unique()## [5.0, 8.0, 11.0, 15.0, 13.0, ..., 9.0, 10.0, 14.0, 12.0, NaN]
## Length: 12
## Categories (11, float64): [5.0, 8.0, 11.0, 15.0, ..., 9.0, 10.0, 14.0, 12.0]Some nan but otherwise this is could be a useful feature, I will convert it back into a numerical column so that if we can easily impute the NaN’s based on a mathematical method such as the mean of the column.
df.years_study = df.years_study.astype('interger')## Error in py_call_impl(callable, dots$args, dots$keywords): TypeError: data type 'interger' not understood
##
## Detailed traceback:
## File "<string>", line 1, in <module>
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\generic.py", line 5548, in astype
## new_data = self._mgr.astype(dtype=dtype, copy=copy, errors=errors,)
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\internals\managers.py", line 604, in astype
## return self.apply("astype", dtype=dtype, copy=copy, errors=errors)
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\internals\managers.py", line 409, in apply
## applied = getattr(b, f)(**kwargs)
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\internals\blocks.py", line 548, in astype
## dtype = pandas_dtype(dtype)
## File "C:\Users\bruno\AppData\Local\R-MINI~1\envs\R-RETI~1\lib\site-packages\pandas\core\dtypes\common.py", line 1763, in pandas_dtype
## npdtype = np.dtype(dtype)Another numpy quirk you can’t use integers because there are NaN values.
df.years_study = df.years_study.astype('float')Converting civil_status into a category.
df.civil_status.unique()## array([1., 0.])To know what 1 or 0 mean, so we need to check the dictionary
dict_civil_status = { 0. : "not_married",
1. : "married"}df.civil_status = df.civil_status.replace(dict_civil_status)
df.civil_status.head()## 0 married
## 1 married
## 2 not_married
## 3 married
## 4 married
## Name: civil_status, dtype: objectBefore we deal with numerical variables I will get rid of ‘Unnamed: 0’ and ‘id’ features because they are useless in this case.
df.drop(columns=['Unnamed: 0', 'id'],inplace=True)Seeing the effects of categorical Variables
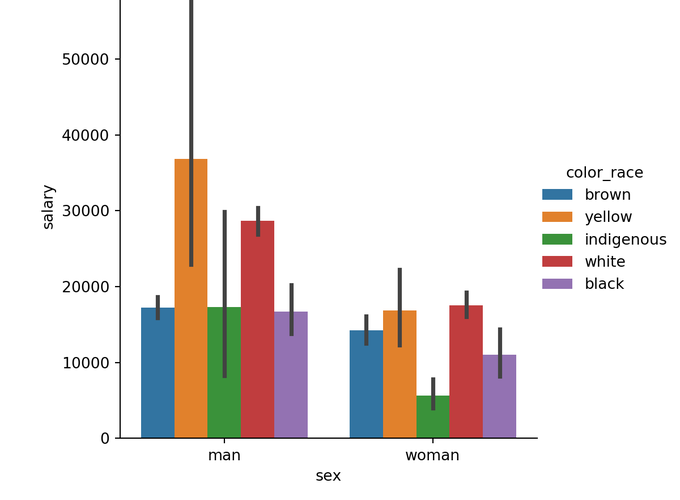
We can use a colored barplot to see the interaction of these Categorical Variables with our Hypothesis.
sns_plot = sns.catplot(x="sex", y="salary", hue="region", kind="bar", data=df)py$sns_plot## <seaborn.axisgrid.FacetGrid>sns_plot = sns.catplot(x="sex", y="salary", hue="civil_status", kind="bar", data=df)
plt.show(sns_plot)
sns_plot = sns.catplot(x="sex", y="salary", hue="color_race", kind="bar", data=df)
plt.show(sns_plot)
Cleaning numerical data
If we pull back the code that we used here are the numerical features of this dataset
df.select_dtypes(include=[np.number]).columns## Index(['age', 'years_study', 'salary'], dtype='object')It is very common to reuse these kind of codes in Data Science scripts, so you shouldn’t fell as bad about repeating yourself as you do in other endeavors such in normal software engendering and you call always clean your analysis latter.
In order to know what to “clean” in numerical data I like to use plot such as a histogram
df.salary.hist(bins = 10)
plt.show()
Here we can see that the data may have a few outliers at 1000000 and that most of the salary data has a large Positive skew meaning that most data point are left to the mean of the dataset we can see that better using an density plot instead
plot_density = df.salary.plot.kde()
plot_density.set_xlim(0,100000)## (0.0, 100000.0)plot_density #### Replacing variables {#python_custom_function_2}
#### Replacing variables {#python_custom_function_2}
If we go back to our custom function we can find that the values -1 and 999999 are unusually common after consulting the dictionary we decided to replace these values with the mean of the group.
This operation would be wrong for machine learning purposes since the mean of our train group would leak information from the test set as well but here in exploratory data analysis it is mostly fine also you need to replace the values with the numpy nan or else this operation doesn’t work as expected.
df_copy = df.copy()
df_copy.salary.replace({-1: "NaN",999999:'NaN'},inplace = True)
df_copy.salary.fillna(df.salary.mean(),inplace= True)pretty_value_counts(pd.DataFrame(df_copy.salary))## Column salary with object data type
## counts percentages cum_percentages
## 19706.790323432902 18592 0.279705 0.279705
## 0.0 1841 0.027697 0.307402
## NaN 1468 0.022085 0.329487
## 5229.0 277 0.004167 0.333654
## 7200.0 260 0.003912 0.337566# Create the new na values
df.salary.replace({-1:np.nan,999999:np.nan},inplace = True)
df.salary.fillna(df.salary.mean(),inplace= True)pretty_value_counts(pd.DataFrame(df.salary))## Column salary with float64 data type
## counts percentages cum_percentages
## 12422.39119 20060 0.301790 0.301790
## 0.00000 1841 0.027697 0.329487
## 5229.00000 277 0.004167 0.333654
## 7200.00000 260 0.003912 0.337566
## 7560.00000 244 0.003671 0.341237And that is the magic of mutable Data Structures no extra assignments are required, quite useful, but be careful there is no going back if you haven’t saved a copy of your data.
Log of numerical data
There is also a statisticall solution for the Positive skew in our Data we can take the log of the salary column, but we will have to add one to all values since log of 0 goes to -Inf
df.log_salary = np.log1p(df.salary)## C:/Users/bruno/AppData/Local/r-miniconda/envs/r-reticulate/python.exe:1: UserWarning: Pandas doesn't allow columns to be created via a new attribute name - see https://pandas.pydata.org/pandas-docs/stable/indexing.html#attribute-accesspretty_value_counts(pd.DataFrame(df.log_salary))## Column salary with float64 data type
## counts percentages cum_percentages
## 9.427336 20060 0.301790 0.301790
## 0.000000 1841 0.027697 0.329487
## 8.562167 277 0.004167 0.333654
## 8.881975 260 0.003912 0.337566
## 8.930759 244 0.003671 0.341237But then it is gonne
df.info()## <class 'pandas.core.frame.DataFrame'>
## RangeIndex: 66470 entries, 0 to 66469
## Data columns (total 7 columns):
## # Column Non-Null Count Dtype
## --- ------ -------------- -----
## 0 age 66470 non-null int64
## 1 sex 66470 non-null object
## 2 years_study 66036 non-null float64
## 3 color_race 66228 non-null object
## 4 salary 66470 non-null float64
## 5 civil_status 66470 non-null object
## 6 region 66470 non-null object
## dtypes: float64(2), int64(1), object(4)
## memory usage: 3.6+ MBYou are better off using the [ notation
df['log_salary'] = np.log1p(df.salary)plot_density = df.log_salary.plot.kde(bw_method= 0.5)
plot_density.set_xlim(0,15)## (0.0, 15.0)plot_density
It is now a usefull feature for most simple linear models
Other numerical columns
df.age.hist(bins = 20)
plt.show()
plot_density = df.age.plot.kde()
plot_density
pretty_value_counts(pd.DataFrame(df.age))## Column age with int64 data type
## counts percentages cum_percentages
## 20 2104 0.031653 0.031653
## 28 2056 0.030931 0.062585
## 26 2040 0.030691 0.093275
## 22 2034 0.030600 0.123875
## 27 2017 0.030345 0.154220Age seems fine
Remember from the the categorical variables we passed years_study here so that we could impute its missing values
df.info()## <class 'pandas.core.frame.DataFrame'>
## RangeIndex: 66470 entries, 0 to 66469
## Data columns (total 8 columns):
## # Column Non-Null Count Dtype
## --- ------ -------------- -----
## 0 age 66470 non-null int64
## 1 sex 66470 non-null object
## 2 years_study 66036 non-null float64
## 3 color_race 66228 non-null object
## 4 salary 66470 non-null float64
## 5 civil_status 66470 non-null object
## 6 region 66470 non-null object
## 7 log_salary 66470 non-null float64
## dtypes: float64(3), int64(1), object(4)
## memory usage: 4.1+ MBWe are missing 66470 - 66036 = 434 observation, this is a small enough number that we decided to drop these rows
While we are droping missing values lets drop the color_race missing observations as well
df.dropna(subset = ["years_study","color_race"],inplace= True)df.info()## <class 'pandas.core.frame.DataFrame'>
## Int64Index: 65795 entries, 0 to 66469
## Data columns (total 8 columns):
## # Column Non-Null Count Dtype
## --- ------ -------------- -----
## 0 age 65795 non-null int64
## 1 sex 65795 non-null object
## 2 years_study 65795 non-null float64
## 3 color_race 65795 non-null object
## 4 salary 65795 non-null float64
## 5 civil_status 65795 non-null object
## 6 region 65795 non-null object
## 7 log_salary 65795 non-null float64
## dtypes: float64(3), int64(1), object(4)
## memory usage: 4.5+ MBChecking on year_study
df.years_study.hist(bins = 20)
plt.show()
Let’s convert it back into a Category
df.years_study = df.years_study.astype('category')Saving our work for later
Here we have many options we can for example run this script later or save this modified df as a csv, both options are okay but I will promote the usage of an Data format that keeps the mindful choices of encoding that we made into consideration, there are many alternatives in this case as well but I will use feather.
It is also always a good idea to separate the Data from the script if you want reproducible work, that is where Excel mostly fails for me.
So showing our Data Types
df.dtypes## age int64
## sex object
## years_study category
## color_race object
## salary float64
## civil_status object
## region object
## log_salary float64
## dtype: objectUsing csv will may lose some Data Types
df.to_csv(file_path_linux + '/finished_work.csv')pd.read_csv(file_path_linux + '/finished_work.csv').dtypes## Unnamed: 0 int64
## age int64
## sex object
## years_study float64
## color_race object
## salary float64
## civil_status object
## region object
## log_salary float64
## dtype: objectWe lost our encoding of years_study and when writing a csv we made this useless to us Unnamed: 0 column
a better way is using the feather file format, you need to pip install pyarrow beforehand
df.reset_index().to_feather(file_path_linux + '/sex_thesis_assignment.feather')pd.read_feather(file_path_linux + '/sex_thesis_assignment.feather').dtypes## index int64
## age int64
## sex object
## years_study category
## color_race object
## salary float64
## civil_status object
## region object
## log_salary float64
## dtype: objectFeather does keep the years study dtype, but feather is still in a experimental phase so be carefull with it, parquet unfortunally fails to keep the dtypes I don’t know why.
It is also a good idea to keep good file names so that you can easily identify your datasets and scripts.
If you then need to delete these files you can do it inside python
os.remove(file_path_linux +'/finished_work.csv')
#os.remove(file_path_linux + '/sex_thesis_assignment.feather')Next post
In the next post I will show the end of the analysis and the “answer” to our hypothesis.